GPU non-cartesian reconstruction with SENSE
Contents
Note
Click here to download the full example code or to run this example in your browser via Binder
GPU non-cartesian reconstruction with SENSE#
Author: Chaithya G R
In this tutorial we will reconstruct an MR Image directly with density compensation and SENSE from gpuNUFFT
Import neuroimaging data#
We use the toy datasets available in pysap, more specifically a 3D orange data and the radial acquisition scheme (non-cartesian).
Package import
from mri.operators import NonCartesianFFT, WaveletN
from mri.operators.utils import normalize_frequency_locations
from mri.operators.fourier.utils import estimate_density_compensation
from mri.reconstructors import SelfCalibrationReconstructor
from mri.reconstructors.utils.extract_sensitivity_maps import get_Smaps
from pysap.data import get_sample_data
# Third party import
from modopt.math.metrics import ssim
from modopt.opt.linear import Identity
from modopt.opt.proximity import SparseThreshold
import numpy as np
import matplotlib.pyplot as plt
Loading input data
image = get_sample_data('3d-pmri').data.astype(np.complex64)
cartesian = np.linalg.norm(image, axis=0)
# Obtain MRI non-cartesian mask and estimate the density compensation
radial_mask = get_sample_data("mri-radial-3d-samples")
kspace_loc = normalize_frequency_locations(radial_mask.data)
density_comp = estimate_density_compensation(kspace_loc, cartesian.shape, 'pipe', backend='gpunufft')
Out:
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mri/operators/fourier/utils/processing.py:101: UserWarning: Frequencies outside the 0.5 limit.
warnings.warn("Frequencies outside the 0.5 limit.")
View Input
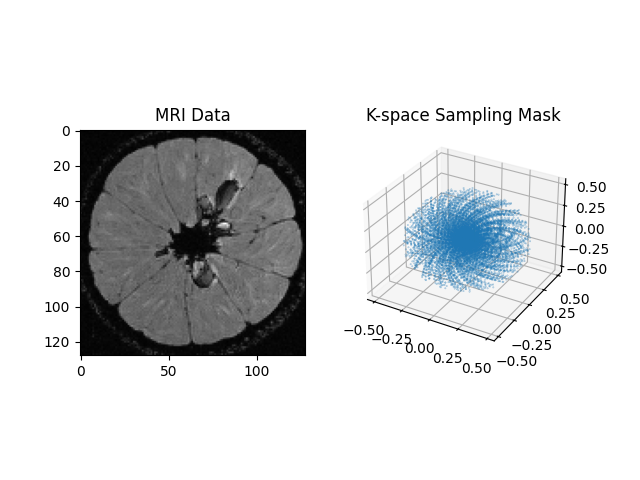
plt.subplot(1, 2, 1)
plt.imshow(cartesian[..., 80], cmap='gray')
plt.title("MRI Data")
ax = plt.subplot(1, 2, 2, projection='3d')
ax.scatter(*kspace_loc[::500].T, s=0.1, alpha=0.5)
plt.title("K-space Sampling Mask")
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_gpuNUFFT_SENSE_DensityCompensation.py:51: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Generate the kspace#
From the 3D orange slice and 3D radial acquisition mask, we retrospectively undersample the k-space We then reconstruct using adjoint with and without density compensation
# Get the locations of the kspace samples and the associated observations
fourier_op = NonCartesianFFT(
samples=kspace_loc,
shape=cartesian.shape,
n_coils=image.shape[0],
implementation='gpuNUFFT',
)
kspace_obs = fourier_op.op(image)
Obtrain the Sensitivity Maps
Smaps, SOS = get_Smaps(
k_space=kspace_obs,
img_shape=fourier_op.shape,
samples=kspace_loc,
thresh=(0.05, 0.05, 0.05), # The cutoff threshold in each kspace
# direction between 0 and kspace_max (0.5)
min_samples=kspace_loc.min(axis=0),
max_samples=kspace_loc.max(axis=0),
density_comp=density_comp,
mode='NFFT',
)
Out:
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/_utils.py:36: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/operators/base.py:434: UserWarning: coeffs should be of dtype complex128. Casting it for you.
warnings.warn(
View Input
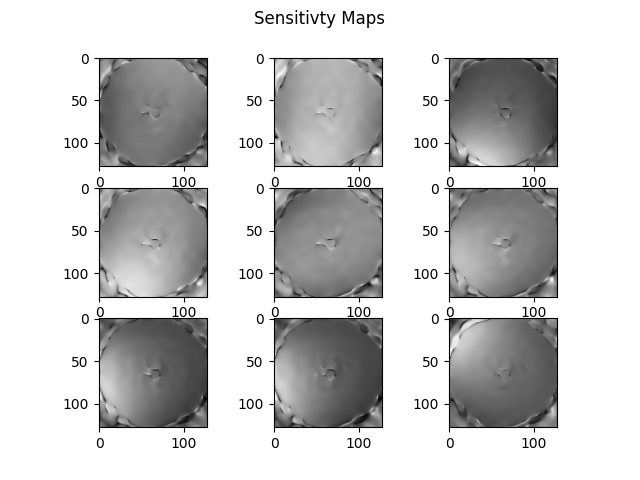
for i in range(9):
plt.subplot(3, 3, i+1)
plt.imshow(np.abs(Smaps[i][..., 80]), cmap='gray')
plt.suptitle("Sensitivty Maps")
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_gpuNUFFT_SENSE_DensityCompensation.py:88: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Density Compensation adjoint:
fourier_op_sense_dc = NonCartesianFFT(
samples=kspace_loc,
shape=cartesian.shape,
implementation='gpuNUFFT',
n_coils=image.shape[0],
density_comp=density_comp,
smaps=Smaps,
)
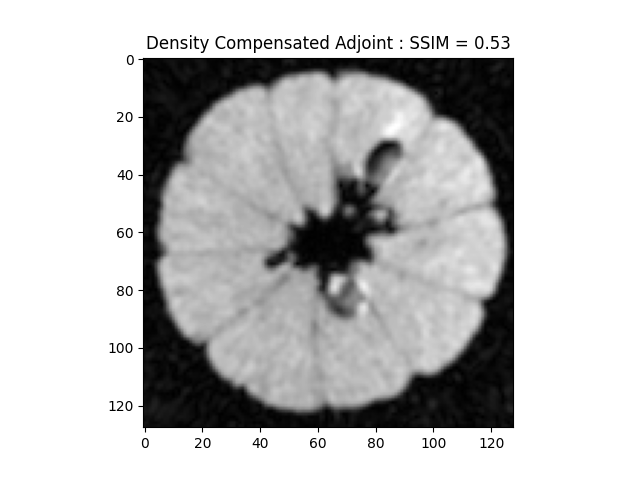
# This preconditions k-space giving a result closer to inverse
image_rec = fourier_op_sense_dc.adj_op(kspace_obs)
recon_ssim = ssim(image_rec, cartesian, mask=np.abs(image)>np.mean(np.abs(image)))
plt.imshow(np.abs(image_rec)[..., 80], cmap='gray')
plt.title('Density Compensated Adjoint : SSIM = ' + str(np.around(recon_ssim, 2)))
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/operators/interfaces/gpunufft.py:138: UserWarning: no pinning provided, pinning existing smaps now.
warnings.warn("no pinning provided, pinning existing smaps now.")
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_gpuNUFFT_SENSE_DensityCompensation.py:105: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
FISTA optimization#
We now want to refine the zero order solution using a FISTA optimization. The cost function is set to Proximity Cost + Gradient Cost
# Setup the operators
linear_op = WaveletN(
wavelet_name='sym8',
nb_scale=4,
dim=3,
)
regularizer_op = SparseThreshold(Identity(), 1e-11, thresh_type="soft")
# Setup Reconstructor
reconstructor = SelfCalibrationReconstructor(
fourier_op=fourier_op_sense_dc,
linear_op=linear_op,
regularizer_op=regularizer_op,
gradient_formulation='synthesis',
verbose=1,
)
Out:
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/pywt/_multilevel.py:43: UserWarning: Level value of 4 is too high: all coefficients will experience boundary effects.
warnings.warn(
WARNING: Making input data immutable.
Lipschitz constant is 0.3916741609573365
The lipschitz constraint is satisfied
Run the FISTA reconstruction and view results
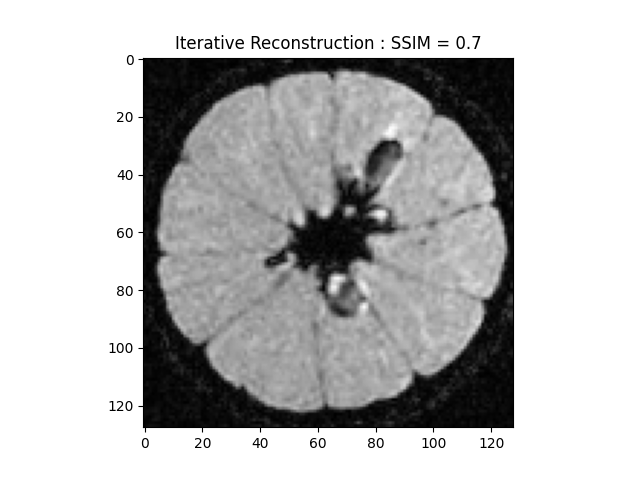
image_rec, costs, metrics = reconstructor.reconstruct(
kspace_data=kspace_obs,
optimization_alg='fista',
num_iterations=30,
)
recon_ssim = ssim(image_rec, cartesian)
plt.imshow(np.abs(image_rec)[..., 80], cmap='gray')
plt.title('Iterative Reconstruction : SSIM = ' + str(np.around(recon_ssim, 2)))
plt.show()
Out:
WARNING: Making input data immutable.
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/pywt/_multilevel.py:43: UserWarning: Level value of 4 is too high: all coefficients will experience boundary effects.
warnings.warn(
- mu: 1e-11
- lipschitz constant: 0.3916741609573365
- data: (128, 128, 160)
- wavelet: <mri.operators.linear.wavelet.WaveletN object at 0x7f3a681ca0e0> - 4
- max iterations: 30
- image variable shape: (128, 128, 160)
- alpha variable shape: (4017261,)
----------------------------------------
Starting optimization...
0%| | 0/30 [00:00<?, ?it/s]
3%|3 | 1/30 [00:03<01:39, 3.42s/it]
7%|6 | 2/30 [00:06<01:35, 3.41s/it]
10%|# | 3/30 [00:10<01:31, 3.40s/it]
13%|#3 | 4/30 [00:13<01:28, 3.40s/it]
17%|#6 | 5/30 [00:17<01:25, 3.40s/it]
20%|## | 6/30 [00:20<01:21, 3.40s/it]
23%|##3 | 7/30 [00:23<01:18, 3.41s/it]
27%|##6 | 8/30 [00:27<01:14, 3.41s/it]
30%|### | 9/30 [00:30<01:11, 3.41s/it]
33%|###3 | 10/30 [00:34<01:08, 3.41s/it]
37%|###6 | 11/30 [00:37<01:04, 3.41s/it]
40%|#### | 12/30 [00:40<01:01, 3.41s/it]
43%|####3 | 13/30 [00:44<00:57, 3.41s/it]
47%|####6 | 14/30 [00:47<00:54, 3.41s/it]
50%|##### | 15/30 [00:51<00:51, 3.41s/it]
53%|#####3 | 16/30 [00:54<00:47, 3.41s/it]
57%|#####6 | 17/30 [00:57<00:44, 3.41s/it]
60%|###### | 18/30 [01:01<00:40, 3.41s/it]
63%|######3 | 19/30 [01:04<00:37, 3.40s/it]
67%|######6 | 20/30 [01:08<00:34, 3.41s/it]
70%|####### | 21/30 [01:11<00:30, 3.41s/it]
73%|#######3 | 22/30 [01:14<00:27, 3.41s/it]
77%|#######6 | 23/30 [01:18<00:23, 3.41s/it]
80%|######## | 24/30 [01:21<00:20, 3.41s/it]
83%|########3 | 25/30 [01:25<00:17, 3.40s/it]
87%|########6 | 26/30 [01:28<00:13, 3.39s/it]
90%|######### | 27/30 [01:31<00:10, 3.39s/it]
93%|#########3| 28/30 [01:35<00:06, 3.39s/it]
97%|#########6| 29/30 [01:38<00:03, 3.39s/it]
100%|##########| 30/30 [01:42<00:00, 3.39s/it]
100%|##########| 30/30 [01:42<00:00, 3.40s/it]
- final iteration number: 30
- final log10 cost value: 6.0
- converged: False
Done.
Execution time: 102.08925738791004 seconds
----------------------------------------
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_gpuNUFFT_SENSE_DensityCompensation.py:140: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Total running time of the script: ( 3 minutes 49.593 seconds)
