GPU Density Compensation Reconstruction Comparison
Contents
Note
Click here to download the full example code or to run this example in your browser via Binder
GPU Density Compensation Reconstruction Comparison#
Author: Chaithya G R
In this tutorial we will reconstruct an MR Image directly with density compensation
Import neuroimaging data#
We use the toy datasets available in pysap, more specifically a 2D brain slice and the radial acquisition scheme (non-cartesian).
Package import
from mri.operators import NonCartesianFFT
from mri.operators.fourier.utils import estimate_density_compensation, \
convert_locations_to_mask, gridded_inverse_fourier_transform_nd
from pysap.data import get_sample_data
# Third party import
from modopt.math.metrics import ssim
import numpy as np
import matplotlib.pyplot as plt
Loading input data
image = get_sample_data('2d-mri').data.astype(np.complex64)
# Obtain MRI non-cartesian mask and estimate the density compensation
kspace_loc = get_sample_data("mri-radial-samples").data
density_comp = estimate_density_compensation(kspace_loc, image.shape, 'pipe', backend='gpunufft')
density_comp2 = estimate_density_compensation(kspace_loc, image.shape, 'cell_count')
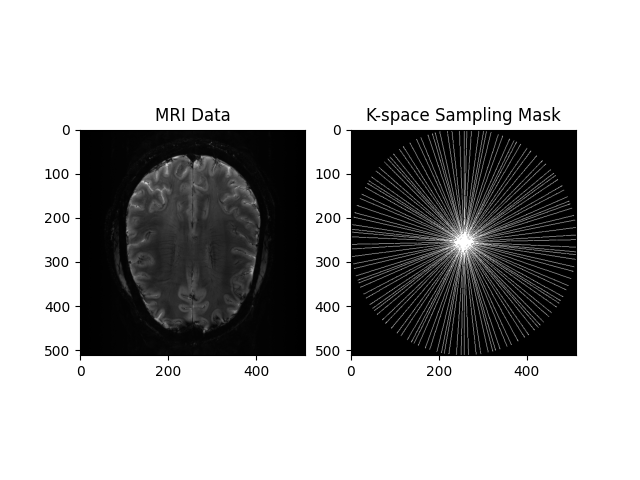
View Input
plt.subplot(1, 2, 1)
plt.imshow(np.abs(image), cmap='gray')
plt.title("MRI Data")
plt.subplot(1, 2, 2)
plt.imshow(convert_locations_to_mask(kspace_loc, image.shape), cmap='gray')
plt.title("K-space Sampling Mask")
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_Pipe_DensityCompensation.py:45: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Generate the kspace#
From the 2D brain slice and the acquisition mask, we retrospectively undersample the k-space using a radial acquisition mask We then reconstruct using adjoint with and without density compensation
# Get the locations of the kspace samples and the associated observations
fourier_op = NonCartesianFFT(
samples=kspace_loc,
shape=image.shape,
)
fourier_op_density_comp = NonCartesianFFT(
samples=kspace_loc,
shape=image.shape,
density_comp=density_comp
)
fourier_op_density_comp2 = NonCartesianFFT(
samples=kspace_loc,
shape=image.shape,
density_comp=density_comp2
)
# Get the kspace data retrospectively. Note that this can be done with
# `fourier_op_density_comp` as the forward operator is the same
kspace_obs = fourier_op.op(image)
Out:
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/_utils.py:36: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/_utils.py:36: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/Chaithya/actions-runner/_work/_tool/Python/3.10.12/x64/lib/python3.10/site-packages/mrinufft/_utils.py:36: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
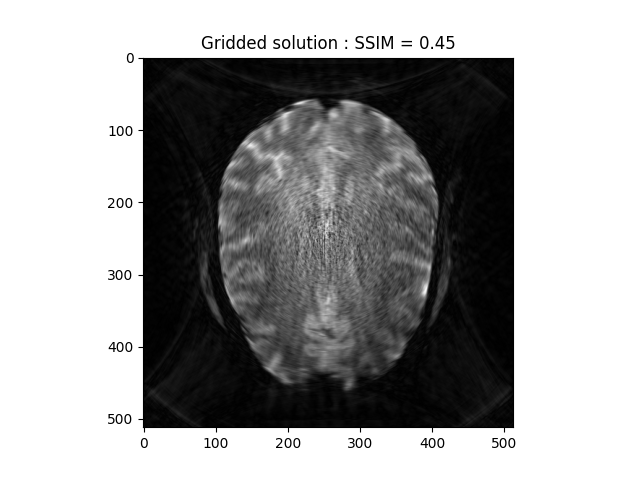
Gridded solution
grid_space = np.linspace(-0.5, 0.5, num=image.shape[0])
grid2D = np.meshgrid(grid_space, grid_space)
grid_soln = gridded_inverse_fourier_transform_nd(kspace_loc, kspace_obs,
tuple(grid2D), 'linear')
base_ssim = ssim(grid_soln, image, mask=np.abs(image)>np.mean(np.abs(image)))
plt.imshow(np.abs(grid_soln), cmap='gray')
plt.title('Gridded solution : SSIM = ' + str(np.around(base_ssim, 2)))
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_Pipe_DensityCompensation.py:83: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Simple adjoint This preconditions k-space giving a result closer to inverse
image_rec = fourier_op.adj_op(kspace_obs)
recon_ssim = ssim(image_rec, image, mask=np.abs(image)>np.mean(np.abs(image)))
plt.imshow(np.abs(image_rec), cmap='gray')
plt.title('Simple NUFFT Adjoint : SSIM = ' + str(np.around(recon_ssim, 2)))
plt.show()

Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_Pipe_DensityCompensation.py:91: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
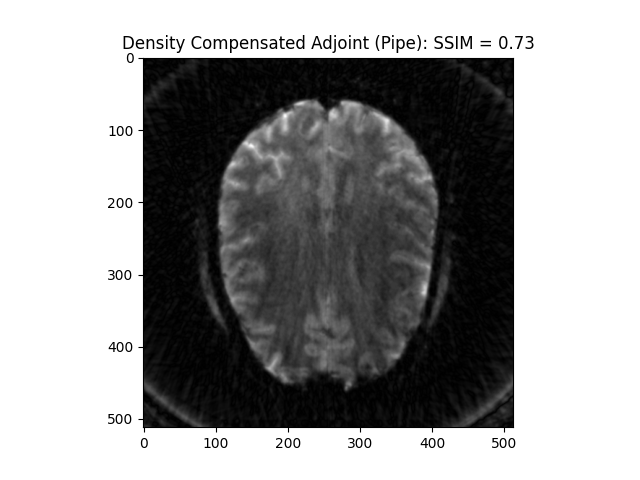
Density Compensation adjoint (from Pipe): This preconditions k-space giving a result closer to inverse
image_rec = fourier_op_density_comp.adj_op(kspace_obs)
recon_ssim = ssim(image_rec, image, mask=np.abs(image)>np.mean(np.abs(image)))
plt.imshow(np.abs(image_rec), cmap='gray')
plt.title('Density Compensated Adjoint (Pipe): SSIM = ' + str(np.around(recon_ssim, 2)))
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_Pipe_DensityCompensation.py:100: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
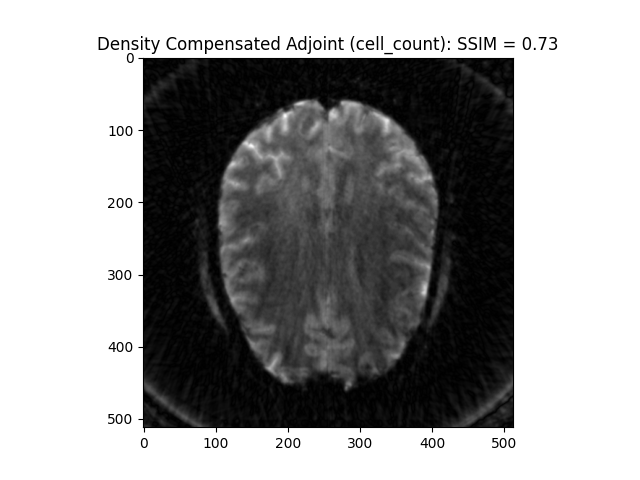
Density Compensation adjoint (from cell_count): This preconditions k-space giving a result closer to inverse
image_rec = fourier_op_density_comp.adj_op(kspace_obs)
recon_ssim = ssim(image_rec, image, mask=np.abs(image)>np.mean(np.abs(image)))
plt.imshow(np.abs(image_rec), cmap='gray')
plt.title('Density Compensated Adjoint (cell_count): SSIM = ' + str(np.around(recon_ssim, 2)))
plt.show()
Out:
/volatile/Chaithya/actions-runner/_work/pysap/pysap/examples/pysap-mri/GPU_NonCartesian_Pipe_DensityCompensation.py:109: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
plt.show()
Total running time of the script: ( 0 minutes 3.200 seconds)
